
|
In this section, have been grouped, the software used to build the primers
using the Nearest-Neighbor thermodynamic parameters
|
-
Human Genomic PickPrimer (with variant)
(create PCR Primers on regions without variants reported in various DBs)
-
Sequence PickPrime
(pick primers from a DNA sequence)
This section contains the software that analyze PCR products (amplicons)
and the software that analyze the oligonucleotide sequences and the primers sequences
-
Amplicon Analysis
(Analysis Template and Primer secondary structure in PCR reaction)
-
Oligo Melting
(Melting temperature and Thermodynamic Details of DNA Hybridization)
-
PCR Primer Analysis
(hairpin-loop, dimer, bases penality and melting temperature)
-
Sequencing Primer Analysis
(Analysis of primers used for Sanger sequencing)
-
Primer Multiplex PCR
(Analyze formations of structures (dimers and hairpin-loops) between different primers used in multiplex reactions)
 |
In this section are grouped the software related
to the sequencing with the Sanger methods
(analysis of primers and chromatograms)
|
-
Primer Sequencing Analysis
(Analysis of primers used for Sanger sequencing)
-
ABI Chromatogram

(online ABI sequence Chromatogram)
-
Chromatogram Align
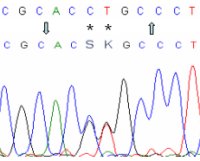
(Align multiple chromatograms with a polymorphic reference sequence)
In this section there are various tools that help manipulate nucleotide sequences or create PCR reactions
This section contains applications related to the human genome and mRNA
-
determines the chromosomal position from one genomic release to another (hg19, hg38 and hs1 )
-
Human Genomics Sequence (hg19/hg38)(with variants)
(returns the sequence of a human genomic region with colored polymorphisms as a function of frequency)
-
Human Exones Gene (new release 2025_12)
shows the position of the exons and the coding sequence
-
Human Align Transcript (with variants) (new release 2025_07)
Aligns mRNA sequences (with variants) with genomic (cDNA) and protein
New: link with HGVS_easy
This section contains applications for analyzing genomic variants
HGVS easy New release 2025_09
(utility to easily write variants in HGVS format and pre-normalize them)
Shows details of a human variant such as the biological effect, isoforms, any related diseases and Pubmed citations.
Converts the descriptions of the human variants (genomics, mRNA and protein) in the forms HGVS, VCF and local.
Appliation for analyze HLA alleles sequence
PriorVar (under testing) ");
(Management of genomic variants from human exome sequencing)
(Prioritization of genomic variants, contained in a VCF file, using functional consequence,
customizable pathogenicity rancking score and LSDB Disease (currently only ClinVar))
|

